ABSTRACT:
In this article, we will discuss about the fascinating world of whole genome assembly. The human genome, comprising over 3 billion base pairs of DNA, holds the blueprint of life itself. Deciphering this intricate code has been one of the most monumental achievements in scientific history. It is a process akin to piecing together a complex jigsaw puzzle, plays a pivotal role in this endeavor. We will also provide references to learn and acknowledge the concept of revealing genetic code.
INTRODUCTION-WHOLE GENOME ASSEMBLY:
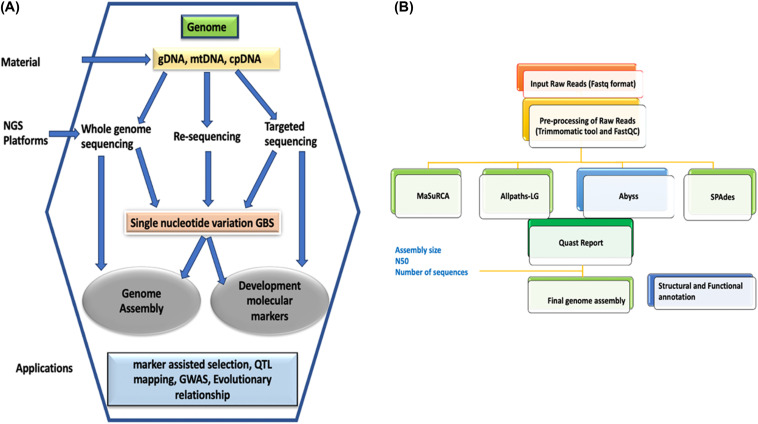
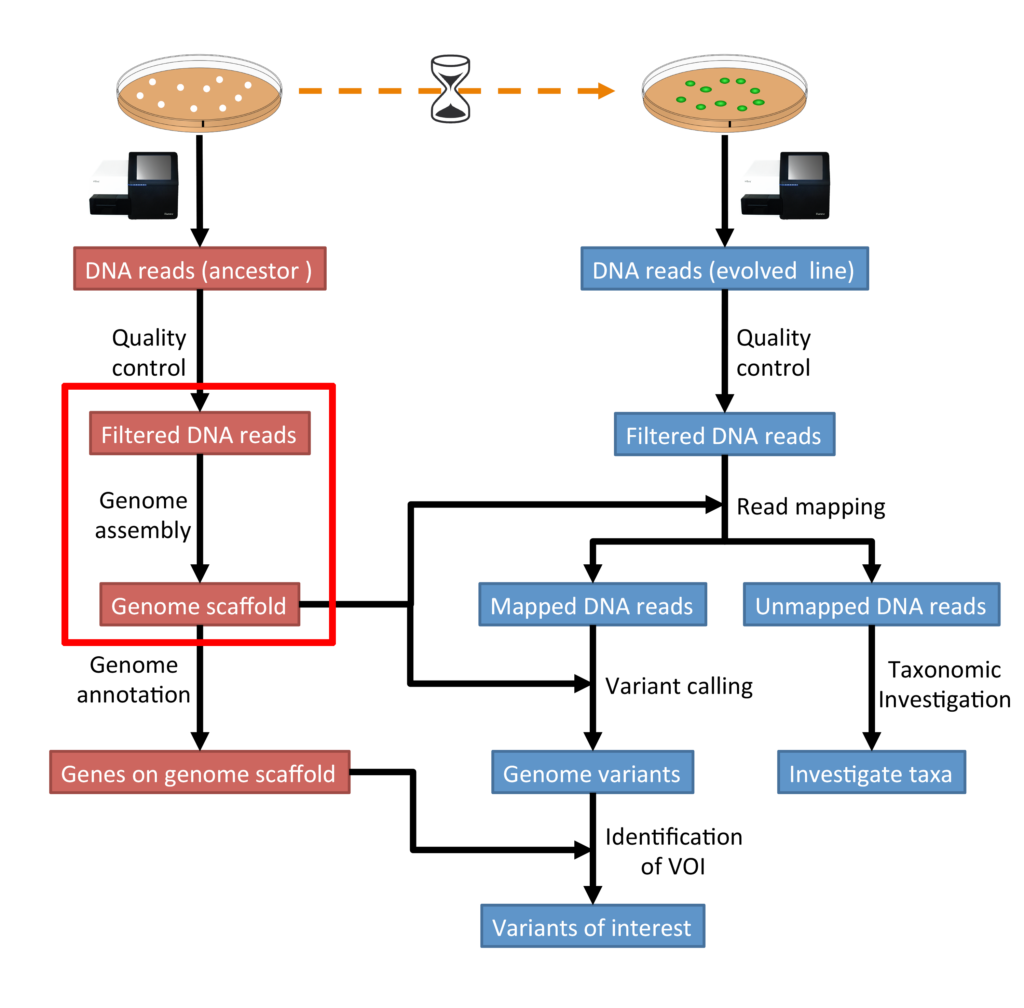
The process of recreating an organism’s whole genome from brief DNA sequences produced by high-throughput sequencing methods known as whole genome assembly. These sequences, referred to as reads, each offer a snapshot of a tiny section of the genome, much like the dispersed parts of a puzzle. Scientists can recreate the full genome sequence, exposing its structure, organization, and genetic variants, by aligning and overlapping these reads.
PROCEDURE OF WHOLE GENOME ASSEMBLY:
1. DATA GENERATION:
Short DNA sequences are first created using older technologies like shotgun sequencing or more recent ones like nanopore sequencing and single-molecule real-time (SMRT) sequencing.
2. PREPROCESSING OF DATA:
To eliminate adaptor sequences, low-quality reads, and other artifacts that can impede the assembly process, raw sequence data preprocessed.
3. ASSEMBLY ALGORITHM:
Contiguous sequences, or contigs, created by assembling the reads using a variety of software tools and algorithms. These algorithms solve the challenging problem of reconstructing the genome by utilizing several techniques, such as overlap-layout-consensus (OLC) and de Bruijn graph-based methods.
4. CONTIGS EXTENSION AND SCAFFOLDING:
Scaffolds are larger structures made up of contigs that are expanded and structured based on additional information like paired-end reads or long-range mapping data. This stage enhances the precision and continuity of the assembly by filling in the spaces between contigs.
5. QUALITY ASSESSMENT:
A thorough quality assessment is performed on the assembled genome to determine its accuracy, continuity, and completeness. Metrics that reveal information about the quality of the assembly include mis-assembly rate, genome coverage, and N50 length.
6. ANNOTATION:
The genome is annotated to identify genes, regulatory elements, and other functional elements after the assembly is judged sufficient. This process offers important insights into the organism’s genetic makeup.
APPLICATIONS-WHOLE GENOME ASSEMBLY:
The study of whole genome assembly holds significant implications not only in genetics but also in evolutionary biology, agriculture, and medicine.
1. GENOMIC MEDICINES:
Precise assembly of the genome makes it possible to identify mutations that cause disease, which in turn makes personalized medicine methods and targeted therapy development easier.
2. CONSERVATION BIOLOGY:
Understanding a species’ genetic diversity and evolutionary history through genome assembly supports conservation efforts and the management of endangered species.
3. CROP IMPROVEMENT:
Scientific research into crop plants’ genomes has made it possible to find genes linked to desired features, which has sped up breeding efforts for increased resistance, quality, and yield.
CHALLENGES IN WHOLE GENOME ASSEMBLY:
The intricacies of the genome, such as repeated sequences, structural differences, and sequencing errors, make whole genome assembly difficult. Nonetheless, the precision and productivity of the assembly process have significantly increased because to ongoing developments in sequencing technologies, assembly algorithms, and computational resources.
1. LONG READ SEQUENCING TECHNOLOGIES:
Longer reads are produced by technologies like SMRT and nanopore sequencing, which make it easier to assemble complicated sections and lower the mistakes brought on by repetitive sequences.
2. HYBRID APPROACHES:
In order to maximize the benefits of both technologies and increase the precision and consistency of the assembled genome, hybrid assembly techniques integrate data from short-read and long-read sequencing.
3. BIOINFORMATICS TOOLS-WHOLE GENOME ASSEMBLY:
The process of genome assembly has been made more efficient and the accuracy of the assembly results has increased with the advent of specialized bioinformatics tools and algorithms.
CONCLUSION-WHOLE GENOME ASSEMBLY:
A key component of contemporary genomics, whole genome assembly reveals the mysteries contained in the DNA of many creatures. Reconstructing and interpreting genomes with accuracy will surely spark important discoveries and transform our understanding of life itself as long as sequencing technologies and computational approaches continue to progress.
REFERENCES:
Koren, S., & Phillippy, A. M. (2015). One chromosome, one contig: complete microbial genomes from long-read sequencing and assembly. Current opinion in microbiology, 23, 110-120. https://pubmed.ncbi.nlm.nih.gov/25461581/
Rhie, A., McCarthy, S. A., & Koren, S. (2021). Towards complete and error-free genome assemblies of all vertebrate species. Nature, 592(7856), 737-746. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8081667/
Miga, K. H., & Koren, S. (2021). Rhie et al. reply. Nature, 596(7872), E6-E7. https://pubmed.ncbi.nlm.nih.gov/32663838/
Vaser, R., Sović, I., Nagarajan, N., & Šikić, M. (2017). Fast and accurate de novo genome assembly from long uncorrected reads. Genome research, 27(5), 737-746. https://pubmed.ncbi.nlm.nih.gov/28100585/

